
微生物基因组学和生物信息学研究组网站 .
欧竑宇教授长期从事微生物基因组学和生物信息学交叉研究,欧博官网关注耐药条件致病菌和抗生素产生菌链霉菌的比较组学分析,围绕细菌可移动基因组的功能解析开发了一系列生物信息学工具和分子生物学数据库。已在Nucleic Acids Research (7篇)、Journal of Bacteriology、Journal of Molecular Diagnostics、FEBS Letters、Int. J. Biochem. Cell Biol.、PloS Genetics、Molecular Microbiology、PloS ONE等刊物发表三十余篇论文。主持多项863计划和国家自然科学基金项目。入选教育部新世纪优秀人才资助计划和上海市青年科技启明星计划,欧博曾获明治乳业生命科学奖。多次在国内外学术会议上作报告和担任会场主持人,多次担任BMC Genomics、Gene、Biochimie、Journal of Theoretical Biology等十余个国内外生物学刊物的审稿人。
教育经历:
2001.03-2004.02, 博士, 生物信息学, 天津大学理学院, 师从张春霆教授
1998.09-2001.03, 硕士, 发酵工程, 天津科技大学生物工程学院, 师从贾士儒教授
1993.09-1997.07, 本科, 食品工程, 南京农业大学食品科技学院
工作经历:
2012.01- 至今, 上海交通大学,欧博娱乐生命科学技术学院,教授
2006.06-2011.12, 上海交通大学,欧博allbet生命科学技术学院,副教授
2004.06-2006.05, 英国莱斯特大学 (University of Leicester),医学院,博士后研究助理
荣誉奖励/人才计划:
2012/2009年上海交通大学晨星青年学者奖励计划SMC优秀青年教师 (A类); 2010年教育部新世纪优秀人才计划; 2008年明治乳业生命科学奖; 2007年上海市青年科技启明星 (A类); 2006年全国优秀博士学位论文提名论文.
研究兴趣:
1. 肺炎克雷伯菌等多重耐药条件致病菌的比较基因组分析
2. 链霉菌抗生素生物合成基因簇的识别
3. 细菌可移动基因组的生物信息学研究
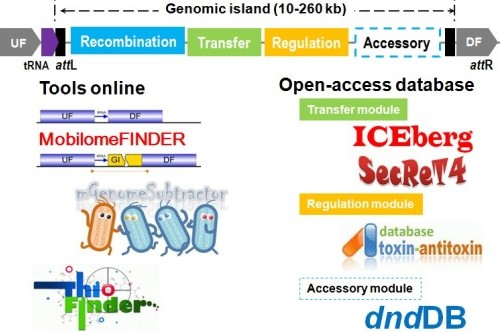
Figure 1. Bioinformatics tools and databased available at the Microbial Genomics and Bioinformatics Group, MML, SJTU.
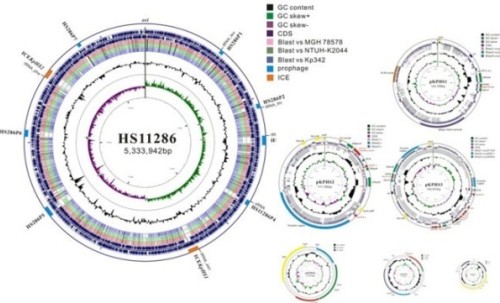
Figure 2. Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain belonging to the dominant clone ST11 of KPC-producing Klebsiella pneumoniae in China. [Liu, et al. Journal of Bacteriology, 2012]
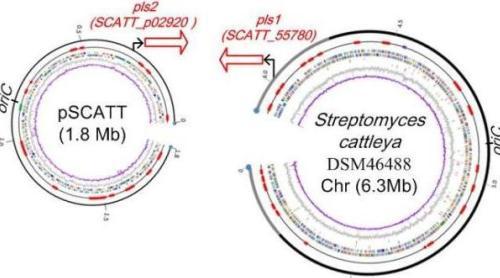
Figure 3. Streptomyces cattleya is unusual for its capacity to biosynthesis of antibiotics and other secondary metabolites, such as fluoroacetate and epsilon-poly-L-Lysine biosynthesis. The mega-plasmid pSCATT is densely packed with 12 putative secondary metabolite gene clusters, in addition to the chromosome that also harbors 17 such gene clusters. [Zhao, et al. Bioorganic Chemistry, 2012]
代表性论文:
1. SecReT4: a web-based bacterial type IV secretion system resource. Bi D, Liu L, Tai C, Deng Z, Rajakumar K*, Ou HY*, Nucleic Acids Research, 2013, 41: D660-D665.
2. Book chapter: “Type II Loci: Phylogeny”. Ou HY*, Wei Y, Bi D, Prokaryotic Toxin-Antitoxins, Editor Kenn Gerdes, 2013, Springer.
3. Bioinformatics tools and databases focused on genomic islands and secondary metabolite biosynthesis of Streptomyces. Ou HY*, Microbiology China, 2013, Actinomycetes issue (in Chinese).
4. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Bi D, Xu Z, Harrison E, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K*, Ou HY*, Nucleic Acids Research, 2012, 40: D621-D626.
5. Complete Genome Sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a Multidrug-Resistant Strain Isolated from Human Sputum. Liu P, Li P, Jiang X, Bi D, Xie Y, Tai C, Deng Z, Rajakumar K, H.Y. Ou*, Journal of Bacteriology, 2012, 194: 1841-1842.
6. ThioFinder: a web-based tool for the identification of thiopeptide gene clusters in DNA sequences. Li J, Qu X, He X, Duan L, Wu G, Bi D, Deng Z, Liu W*, Ou HY*, PloS ONE, 2012, 7: e45878.
7. Insights into fluorometabolite biosynthesis in Streptomyces cattleya DSM46488 through knockout mutants. Zhao C, Li P, Deng Z, Ou HY*, McGlinchey RP, O'Hagan D*, Bioorganic Chemistry, 2012, 44: 1-7.
8. Book chapter: “Identification of bacterial genomic islands by comparative genomic analysis. Ou HY*, Microbiology genomics in China, Editor Zi-Niu Yu, 2011, Science Press (in Chinese).
9. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Shao Y, Harrison EM, Bi D, Tai C, He X, Ou HY*, Rajakumar K, Deng Z, Nucleic Acids Research, 2011, 39: D606-D611.
10. Expansion of the known Klebsiella pneumoniae species gene pool by characterization of novel alien DNA islands integrated into tmRNA gene sites. Zhang J, Jurriaan van Aartsen J, Jiang X, Shao Y, Tai C, He X, Tan Z, Deng Z, Jia S*, Rajakumar K, H.Y. Ou*, Journal of Microbiological Methods, 2011, 84: 283-289.
11. Book chapter: “ArrayOme- & tRNAcc-facilitated mobilome discovery: comparative genomics approaches for identifying rich veins of bacterial novel DNA sequences”. Ou HY, Rajakumar K, Handbook of Molecular Microbial Ecology I: Metagenomics and Complementary Approaches, Editor F. J. de Bruijn, 2011, John Wiley & Sons, Inc.
12. Cleavage of Phosphothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrA. Liu G, Ou HY, Wang T, Li L, Tan H, Zhou X, Rajakumar K, Deng Z*, He X*, PLoS Genetics, 2010, 6: e1001253.
13. mGenomeSubtractor: a web-based tool for parallel in silico subtractive hybridization analysis of multiple bacterial genomes. Shao Y, He X, Tai C, Ou HY*, Rajakumar K, Deng Z, Nucleic Acids Research, 2010, 38: W194-W200.
14. The pheV phenylalanine tRNA gene in Klebsiella pneumoniae clinical isolates is an integration hotspot for possible niche-adaptation genomic islands. Chen N¶, Ou HY¶, van Aartsen JJ, Jiang X, Li M, Yang Z, Wei Q, Chen X, He X, Deng Z, Rajakumar K, Lu Y*, Current Microbiology, 2010, 60, 210-6. (¶ These authors contributed equally to this work.)
15. dndDB: a database focused on phosphorothioation of the DNA backbone. Ou HY, He X, Shao Y, Tai C, Rajakumar K, Deng Z*, PLoS ONE, 2009, 4:e5132.
16. Translational Genomics to Develop a Salmonella enterica Serovar Paratyphi A Multiplex PCR Assay. Ou HY¶, Ju CTS ¶, Thong KL, Ahmad N, Deng Z, Barer MR, Rajakumar K*, Journal of Molecular Diagnostics, 2007, 9: 624-630. (¶ These authors contributed equally to this work.)
17. MobilomeFINDER: web-based tools for in silico and experimental discovery of bacterial genomic islands. Ou HY, He X, Harrison EM, Kulasekara BR, Thani AB, Kadioglu A, Lory S, Hinton JC, Barer MR, Deng Z*, Rajakumar K*, Nucleic Acids Research, 2007, 35, W97-W104.
18. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. He X, Ou HY, Yu Q, Zhou X, Wu J, Liang J, Zhang W, Rajakumar K, Deng Z.*, Molecular Microbiology, 2007, 65, 1034-48.
19. A novel strategy for identification of genomic islands by comparative analysis of the contents and contexts of tRNA sites in closely related bacteria. Ou HY, Chen LL, Lonnen J, Chaudhuri RR, Thani AB, Smith R, Garton NJ, Hinton JC, Pallen M, Barer M, Rajakumar K*, Nucleic Acids Research, 2006, 34: e3.
20. ArrayOme: a program for estimating the sizes of the microarray-visualised genomes. Ou HY, Smith R, Lucchini S, Hinton JC, Chaudhuri RR, Pallen M, Barer M, Rajakumar K*, Nucleic Acids Research, 2005, 33: e3